
CellAtria vs Polly BioAgents is not necessarily an apples to apples comparison as both tools were developed for different use cases. While both leverage agentic AI to streamline bioinformatics, they target different stages of the research lifecycle.
BioAgent is designed as a CLI-first autonomous execution system intended for broad applications across the omics spectrum, including bulk RNA-seq, transcriptomics, and exploratory R&D. It functions as a professional-grade workbench where the agent is responsible for the entire execution loop. Its primary goal is to act as an autonomous extension of a scientific team, allowing researchers to focus on high-level questions while the agent manages the technical heavy lifting of multi-step bioinformatics workflows.
In contrast, CellAtria focuses specifically on the ingestion and standardization of single-cell RNA-seq data. It is built as a dialogue-driven, chatbot-based framework. CellAtria excels at extracting metadata from literature and retrieving datasets from repositories to feed into its assistant pipeline, CellExpress.
To objectively evaluate these systems, two tests were conducted: a "Golden Path" test derived from CellAtria’s supplementary documentation, and a "Stress Test" involving non-standard public data.
This test replicates a primary use case featured in the CellAtria supplementary materials:
“run cellexpress using single cell data from https://www.nature.com/articles/s41467-023-43758-2, no questions asked.”
The Goal: Extract metadata from a specific paper, retrieve the associated data, and run the standard CellExpress/BioAgent pipeline (QC, Doublet Detection, Normalization, HVG, PCA, UMAP, and Leiden Clustering).
To test the limits of autonomy, both agents were tasked with processing GSE96583, a classic single-cell RNA-seq dataset (PBMCs: Control vs. IFN-beta Stimulated) frequently used in bioinformatics tutorials and method papers like the original Seurat and muscat papers because it captures the fundamental challenges of scRNA-seq analysis in a controlled environment. This dataset served as a "stress test" because the public repository lacks standard feature annotation files, requiring the agent to perform complex data engineering to make the data usable.
The Goal: Resolve the relationship between matrix rows and supplementary gene files to reconstruct a valid AnnData object for Differential Expression (DE) analysis.
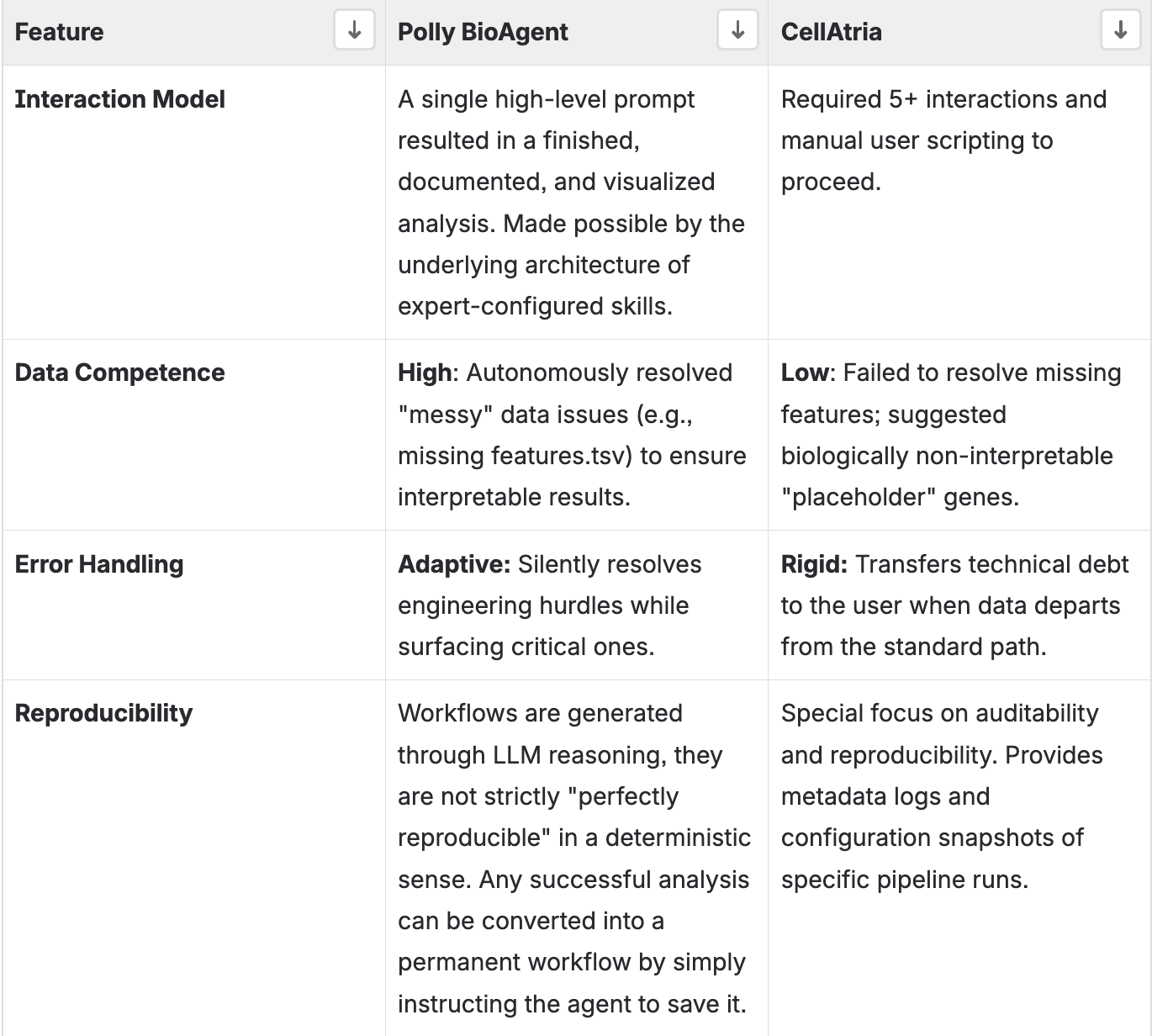
CellAtria is an effective ingestion specialist. It excels at the "Document-to-Metadata" stage, making it a valuable tool for building standardized libraries from published literature. However, it operates as a strict wrapper for the CellExpress pipeline, which limits its flexibility when encountering non-standard data.
Polly BioAgent functions as a partner scientist. It demonstrates the logic required to handle the messiness of exploratory R&D. Its ability to autonomously solve data engineering problems and deliver visualized, biologically valid results in a single interaction makes it superior for end-to-end analysis.