.webp)
The management of advanced prostate cancer (PC) is rapidly evolving with different types of immunotherapies being actively explored. Currently, the only FDA-approved immunotherapy for PC is Sipuleucel-T (Provenge). Though CAR-T therapy has the potential to dramatically change cancer prognosis, extensive research is needed to use it in prostate cancer.
To advance any drug discovery process, an integrative approach is needed which requires analyzing various data types such as transcriptomic, mutation, copy number variation, single-cell data, etc. to find the specific profile for which each medicine would be effective. For eg, Castration-resistant prostate cancer (CRPC) has to be treated differently from other PCs. Though finding and standardizing relevant data is a time-consuming and cumbersome task but it need not always be that way.
The ‘Monthly Dataset Roundup’ series features datasets on Polly that showcase different data types that capture the comprehensive molecular landscape of PC. Well-curated versions of highly relevant datasets from various repositories/sources can be found in a single-step process and analyzed easily using various apps on Polly. These datasets can also be accessed programmatically and analyzed in a Jupyter notebook instance on Polly itself without the need to download the data locally.
Keywords: TCGA, Depmap, Genedependency, LINCS, PharmacoDB, scRNA-Seq
.png)
TCGA Prostate Adenocarcinoma (PRAD) project
Dataset ID: PRAD_*
Year of Publication: 2019
Total Samples: 2434
Experiment type: Transcriptomics, miRNA, CNV, Mutation, Proteomics
Organism: Homo sapiens
Reference link: TCGA-PRAD on GDC


The PRAD project is one of several projects under The Cancer Genome Atlas(TCGA), which aims to study the genotypic-phenotypic relation of major cancers using next-generation sequencing experiments on clinical samples.


L1000 Connectivity Map perturbational profiles from Broad Institute LINCS Center for Transcriptomics LINCS Pilot PHASE I
Dataset ID: lincs_GSE92742_*
Year of Publication: 2017
Total Samples: 35172 (prostate cancer cell line samples) and139,760 (overall samples)
Experiment type: Transcriptomics
Organism: Human cell lines
Reference link: Publication

The Library of Integrated Cellular Signatures (LINCS) is an NIH program that funds the generation of perturbational profiles across multiple cell and perturbation types as well as read-outs at a massive scale. The LINCS Center for Transcriptomics at the Broad Institute uses the L1000 high-throughput gene-expression assay to build a Connectivity Map to enable the discovery of functional connections between drugs, genes, and diseases through the analysis of patterns induced by common changes in gene expression.
In brief, the study design involves the generation of a compendium of transcriptional expression data from cultured human cells treated with small-molecule and genetic loss/gain of function perturbagens. All 35,000+ samples of LINCS studying the effect of perturbagens on prostate cancer cell lines are available on Polly’s LINCS OmixAtlas.



Dose-response data for various small molecules/drug candidates from the Cancer Therapeutics Response Portal version 2 (CTRPv2)generated by the Broad Institute.
Dataset ID: *_prostate_CTRPv2_*
Year of Publication: 2013
Total Samples: 3271 samples for prostate cancer
Experiment type: Dose response
Organism: Human derived cell lines
Reference link: Publication

The Cancer Therapeutics Response Portal (CTRP) was developed by the Center for the Science of Therapeutics at the Broad Institute to screen a large panel of cancer cell lines for sensitivity to small molecules. CTRPv2 is a continuation of the CTRP project and the largest pharmacological screen conducted to date, containing several hundreds of thousands of drug dose-response curves.
The datasets for prostate cancer consist of dose-response (AUC, IC50) data measured for more than 500 drugs, across 6 major prostate cancer cell lines. All of these samples are hosted on the PharmacoDB OmixAtlas on Polly where they can be programmatically accessed and used for downstream applications using the Jupyter notebook environment.

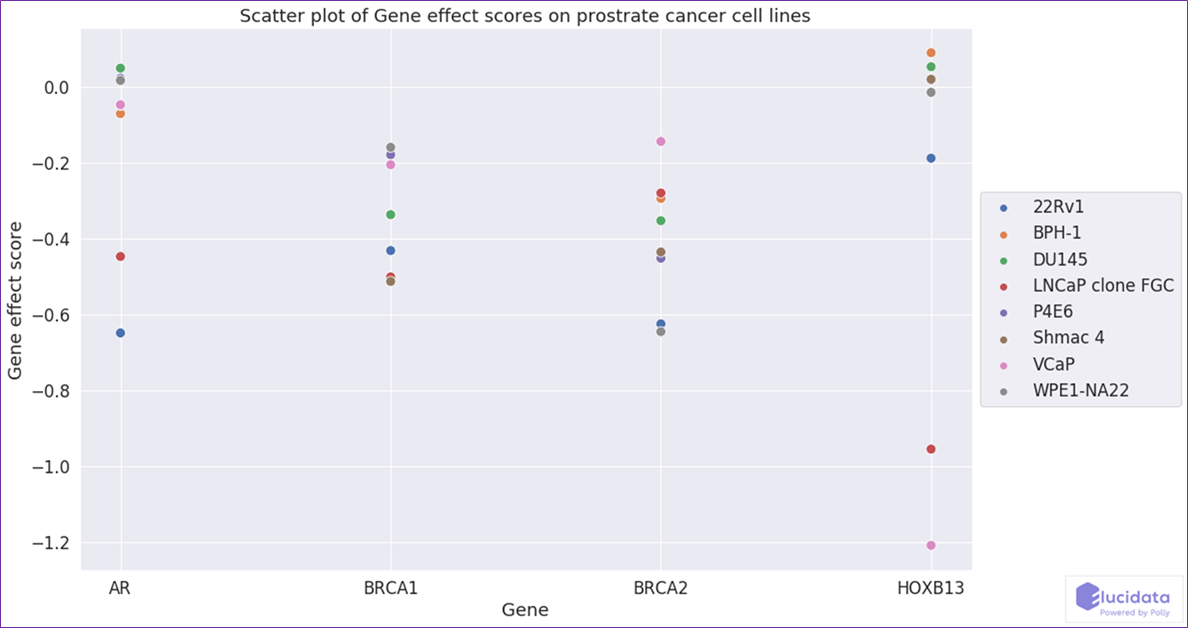
2021 Q4 CRISPR Chronos data from DepMap
Dataset ID: 2021Q4_CRISPR_chronos_gene_dependency_*
Year of Publication: 2021
Total Samples: 1054
Experiment type: Gene dependency
Organism: Human cell lines
Reference link: https://depmap.org/portal/
Developing new cancer therapies is based on finding ways to target processes that will selectively kill cancer cells. The DepMap portal hosts data from genome-wide RNAi and CRISPR loss-of-function screens to systematically identify essential genes across hundreds of human cancers.
CRISPR-Cas9 genetic perturbation reagents are used to silence or knockout individual genes and identify those genes that affect cell survival. By linking these dependencies to the genetic or molecular features of the tumors, this project is providing the foundation for the "Cancer Dependency Map”. Data from all quarterly releases of DepMap is curated, standardized, and hosted on the DepMap OmixAtlas on Polly, ready to be used for all ML or bioinformatics applications.

Immunoprofiling of PC infiltrates
Dataset ID: GSE153892_GPL18573
Year of Publication: 2022
Total Samples: 6
Experiment type: Single cell RNASeq
Organism: Homo sapiens, Mus musculus
Reference link: Publication

Tumor-associated macrophages (TAMs) are correlated with the progression of prostatic adenocarcinoma (PCa) but are still poorly described in this context. Here, high-dimensional single-cell RNA-seq was applied to profile the transcriptional landscape of TAMs in PCa. The researchers identified a subset of tumor-infiltrating macrophages that shows a dysregulation in transcriptional pathways associated with lipid metabolism. In human and mouse models of PCa, this subset of macrophages expresses the scavenger receptor MARCO and is characterized by the accumulation of lipid droplets. The study also identified a gene signature derived from MARCO-expressing TAMs that correlates with PCa progression and is associated with shorter disease-free survival. They observed that lipid accumulation in TAMs is promoted by the secretome of cancer cells. Lipid-loading confers to tumor-conditioned macrophages the capability to promote cancer cell migration mediated by CCL6. The findings provide evidence that lipid-loaded TAMs represent a new therapeutic target in PCa.


RNA sequencing of PC and normal tissue from African-Americans and European-Americans
Dataset ID: GSE104131_GPL16791
Year of Publication: 2018
Total Samples: 26
Experiment type: Transcriptomics
Organism: Homo sapiens
Reference link: Publication

The dataset compares transcriptomes of matched tumor-normal samples from African-American and European-American patients
African-American men (AAM) are at a higher risk of dying from the disease compared to European-American men (EAM). The study was conducted to better understand PC molecular diversity that may be underlying these disparities. The researchers ran RNA-sequencing data analysis on high-grade PC to identify genes showing differential tumor versus normal adjacent tissue expression patterns unique to AAM or EAM.
Matched high-grade (GS≥7(4+3)) prostate tumors and adjacent normal specimens from 16 patients (8 AAM and 8 EAM) were subjected to two replicate runs of RNA-sequencing.


Gene expression profiling of treated and untreated primary PC
Dataset ID: GSE102124_GPL17586
Year of Publication: 2018
Total Samples: 22
Experiment type: Trancsriptomics
Organism: Homo sapiens
Reference link: Publication
Primary PC can have extensive microheterogeneity, but its contribution to the later emergence of metastatic castration-resistant prostate cancer (mCRPC) remains unclear. In this study, the researchers micro dissected residual PC foci in radical prostatectomies from 18 men treated with neoadjuvant-intensive androgen deprivation therapy (leuprolide, abiraterone acetate, and prednisone) and analyzed them for resistance mechanisms.
The study showed that neoadjuvant androgen deprivation therapy for PC selects for tumor foci with subclonal genomic alterations, which may comprise the origin of metastatic castration-resistant prostate cancer.

Upregulated PPARG2 facilitates interaction with demethylated AKAP12 gene promoter and suppresses proliferation in PC
Dataset ID: GSE108309_GPL15207
Year of Publication: 2021
Total Samples: 6
Experiment type: Transcriptomics
Organism: Homo sapiens
Reference link: Publication

This study investigates the biological function and molecular mechanism of the nuclear receptor peroxisome proliferator-activated receptor gamma 2(PPARG2) in PC. The results revealed that PPARG2was downregulated in PC, and overexpression of PPARG2 inhibited cell migration, colony formation, invasion, and induced cell cycle arrest of PC cells in vitro. In addition, PPARG2 overexpression modulated the activation of the Aktsignaling pathway, as well as inhibited tumor growth in vivo. Moreover, the mechanistic analysis revealed thatPPARG2 overexpression induced an increased expression level of miR-200b-3p,which targeted 3′ UTR of the downstream targets DNMT3A/3B. It also facilitated interaction with demethylated AKAP12 gene promoter and suppressed cell proliferation in PC.
The results of this study provided the first evidence for a novelPPARG2-AKAP12 axis-mediated epigenetic regulatory network. The study identified a molecular mechanism involving an epigenetic modification that could be possibly targeted as an antitumoral strategy against PC.


Gene expression profiling of glucocorticoid receptor signaling in PC-associated fibroblast cell model (PF179TCAF-shGR-1)
Dataset ID: GSE150432_GPL23126
Year of Publication: 2021
Total Samples: 12
Experiment type: Transcriptomics
Organism: Cell line
Reference link: Publication

Glucocorticoid receptor (GR) has been recently identified as a candidate for acquired anti-androgen and chemotherapy resistance. In this study, cancer-associated fibroblasts(PF179TCAF-shGR-1) were treated for 24h in 4 different treatment groups (1-DMSO=Control treatment, 2 - Dexamthasone, 3 -Doxycycline+Dexamethasone, 4-RU486+Dexamethasone).

Potent stimulation of the androgen receptor instigates a viral mimicryresponse in PC
Dataset ID: GSE187413_GPL18573
Year of Publication: 2022
Total Samples: 9
Experiment type: Transcriptomics
Organism: Cell line
Reference link: Publication
Inhibiting the androgen receptor (AR), a ligand-activated transcription factor, with androgen deprivation therapy is a standard-of-care treatment for metastatic PC. Paradoxically, activation of AR can also inhibit the growth of PC in some patients and experimental systems, but the mechanisms underlying this phenomenon are poorly understood.
This study exploited a potent synthetic androgen, methyltestosterone (MeT),to investigate AR agonist-induced growth inhibition. MeT strongly inhibited the growth of PC cells expressing AR, but not AR-negative models. Genes and pathways regulated by MeT were highly analogous to those regulated by DHT, although MeT induced a quantitatively greater androgenic response in PC cells. MeT potently down-regulated DNA methyltransferases, leading to global DNA hypomethylation. These epigenomic changes were associated with dysregulation of transposable element expression, with long-term MeT treatment resulting in up regulation of endogenous retrovirus (ERV) transcripts. Increased ERV expression led to the accumulation of double-stranded RNA and a “viral mimicry” response characterized by activation of interferon signaling, upregulation of MHC Class I molecules, and enhanced recognition of murine PC cells by CD8+ Tcells.
Positive associations between AR activity and ERVs/anti-viral pathways were evident in patient transcriptomic data, supporting the clinical relevance of the findings. Collectively, the study reveals that the potent androgen MeT can increase the immunogenicity of PC cells via a viral mimicry response. This finding has potential implications for the development of strategies to sensitize this cancer type to immunotherapies.

Polly’s OmixAtlases provide FAIR biomolecular data on the Polly platform enabling researchers to carry out robust data analysis and effective consumption of omics data. Reach out to us at info@elucidata.io for more details.
