-min.jpg)
What if your harmonization engine ran like a precision factory-automated, scalable, and radically cost-efficient rather than a clunky setup sputtering under pressure?
Imagine two assembly lines: one jammed, noisy, and expensive to maintain; the other sleek, streamlined, and optimized to deliver at scale with minimal downtime. Elucidata’s harmonization platform is built on the latter cutting compute costs by 77%, reducing failures by 3x, and delivering 5TB/week in throughput. In a world where every terabyte costs, which line would you choose?
Processing single-cell RNA sequencing (scRNA-seq) data is notoriously resource-intensive and financially burdensome (Kharchenko, 2021; Slovin et al., 2021). These high-resolution datasets demand massive compute and storage, and legacy pipelines often crumble under the weight leading to delays, high failure rates, and runaway cloud bills (Ke et al., 2022).
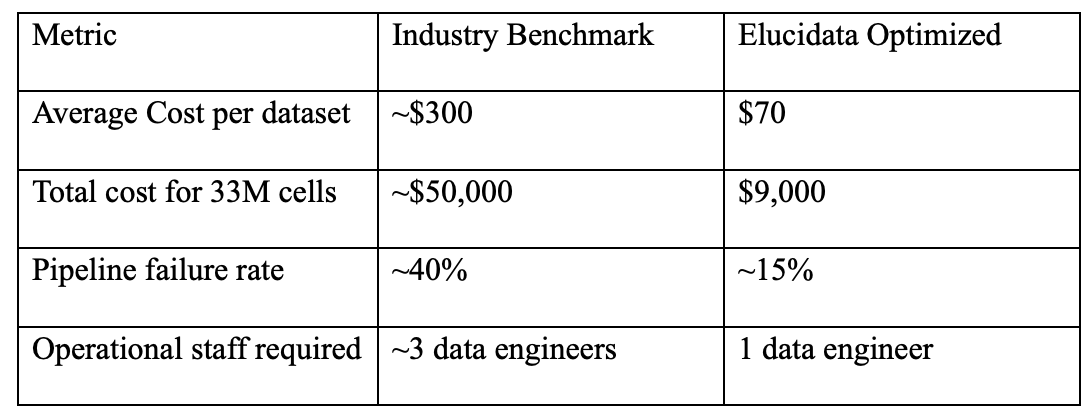
Elucidata faced this challenge firsthand while building its single-cell foundational model: tasked with harmonizing over 33 million single-cell profiles for downstream machine learning. Initial cost projections topped $50,000 even after optimizing with AWS spot instances, making the effort commercially unsustainable without significant infrastructure innovation.
To tackle this, our engineering team rebuilt the harmonization pipeline from the ground up with a singular focus: cut costs without compromising performance, scalability, or stability
The outcome was a lean, scalable harmonization engine that significantly outperformed industry benchmarks across cost, reliability, and throughput.
Infrastructure and Execution Enhancements
Infrastructure Overhaul
The team migrated from AWS Batch to bare-metal clusters, enabling fine-grained control over compute provisioning. This transition eliminated the inefficiency of under-provisioned virtual machines, increasing throughput by 37%. NFS (Network file system) -based scalable shared storage solution was implemented to improve I/O(Input/Output) performance and support concurrent task execution across the pipeline.
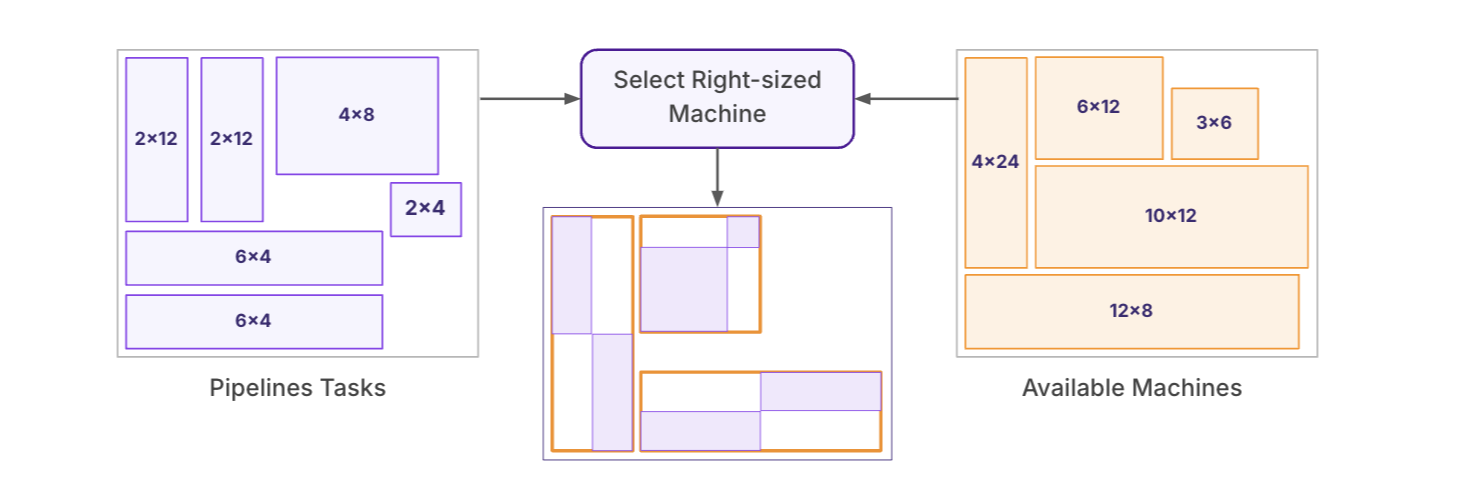
Workflow Cost Optimization
Each stage of the pipeline was analyzed and aligned with its most efficient compute profile using an internal resource-matching algorithm. Adaptive node-packing strategies ensured full utilization of compute nodes. Additionally, a checkpointing mechanism was introduced to cache intermediate data, allowing jobs to resume from the last successful stage, thereby reducing recomputation and cutting compute time and costs by an additional 15%.
Stability and Operational Efficiency
Tool-related instability was tackled head-on by replacing the error-prone faster-dump with the more reliable fastq-dump, significantly reducing decompression failures. A lightweight validation layer was introduced to catch formatting issues, naming inconsistencies, and missing 10x tags before pipeline execution. Centralized dashboards enabled real-time monitoring of task failures and resource utilization, allowing a single engineer to efficiently manage workloads.
Business Impact
These improvements resulted in over $40,000 in cloud compute savings, excluding additional labor cost savings. The redesigned framework now stands as a scalable reference architecture for high-throughput scRNA-seq data harmonization.
Use Cases: Turning Technical Wins into Business Value
The reduced time, cost, and headcount enabled budget previously reserved for troubleshooting and rework enabling faster decision-making in research and development.
Future Implications: A New Playbook for scRNA-seq at Scale
This isn’t just an internal win. The implications stretch across any lab, Contract Research Organisations (CRO), or biopharma company struggling with the high costs of transcriptomics at scale. With our pipeline re-architecture, harmonizing scRNA-seq data becomes accessible, robust, and economically viable even for datasets in the tens of millions.
As AI/ML applications in bioinformatics become more data-hungry (Shandhi & Dunn, 2022) having a harmonization layer that doesn’t bankrupt your compute budget is no longer optional. It’s a strategic necessity.
At Elucidata, we didn’t just cut costs, we redefined what’s possible.
Our reengineered pipeline is more than an internal optimization; it’s a model for scalable, intelligent bioinformatics infrastructure.
The future of single-cell research shouldn’t be limited by budget. It should be defined by what we can discover.
Contact
To explore pilot deployments or assess how this framework can be tailored to your pipeline: info@elucidata.io
References